What's Taking So Long? Code Profiling for Optimization
The guess-and-check approach to code optimization is tempting but faulty- a better me would tell myself to use a profiler, a tool that observes the call stack during the execution of the program to record information, e.g., where the program is spending the most time and how often functions are called. This can also be used to detect bugs, memory leaks (memory profiler), etc.
GNU gprof
The GNU Profiler is the most basic profiler which pairs directly with gcc.
- Compile with
g++ -g -pg [main.cpp]where-gis for debugging and-pgis for the gprof tool. - The code will take longer than normal, and output the executable
a.out. Run the executable to output the result to output a.gmon.outfile - Run the gprof command to interpret the file:
gprof ./a.out gmon.out > test.txt - The default script (above) will create a flat profile, the flag
-qcan be used to create the Call Graph
Code Optimization: Without any optimization flags, the code is very transparent, but not identical to a real run-
this may make some parts of your code take longer, which skews the analysis. Optimization flags like -O1, -O2, -O3
greatly speed up the program, but can also jumble up the code leading to an erroneous analysis. This was observed with
the -O2 flag, where unrelated functions were said to be called. The -O1 flag seems to be a good middle ground for
speed and clarity.
Visualizing the Call Graph:
The call graph gives information about the call stack throughout the program. Below, we see an example: the main()
function is the head of the program, and calls two major functions Hierarchical_HSP_Test() and create_index_2L().
The next largest node is the Hierarchical_HSP_Test() function, and within that, the std::__adjust_heap function
takes the most time. And so forth.
index % time self children called name
[1] 97.6 0.00 18.41 main [1]
6.38 11.14 10000/10000 Hierarchical_HSP_Test() [2]
0.35 0.54 1/1 create_index_2L() [9]
...
-----------------------------------------------
6.38 11.14 10000/10000 main [1]
[2] 92.9 6.38 11.14 10000 Hierarchical_HSP_Test() const [2]
4.87 0.53 15756/15756 std::__adjust_heap [3]
0.18 2.00 35354/35354 validate_point() [5]
1.35 0.40 1497/32112 get_distance() [4]
...
-----------------------------------------------
4.87 0.53 15756/15756 Hierarchical_HSP_Test() [2]
[3] 28.6 4.87 0.53 15756 std::__adjust_heap [3]
0.53 0.00 15756/28609 std::__push_heap [8]
...
While this is simple enough, the call graph above is complex. There are many nodes, the names can be very long, and there can be MANY nodes. This can greatly benefit from a visualization tool.
The gprof2dot library was created for this purpose.
- Given the callgraph outputted to
test.txtby the commandgprof -q ./a.out gmon.out > test.txt - You will need Python3 and the graphviz library
- Download the gprof2dot library, easy way is to install in a virtual
environment with
pip install gprof2dot - To produce an svg file, use the command:
gprof2dot test.txt | dot -Tsvg -o profile.svg. I had an issue with using png or pdf. Can open svg with google chrome.
The gprof2dot library is great. Here are some of the helpful flags:
-nto specify a minimum percentage for nodes to be considered.-eto specify a minimum percentage for edges to be considered.-sto strip the functions of their arguments, templates, etc. Very helpful for long function names-wto wrap function names on seperate lines (more compact)-zto choose a starting root node, rather than themainfunction
Use Cases
Below are some use cases of profiling- it may not be thrilling.
Hierarchical HSP
The Hierarchical HSP (HHSP) is an approach to finding the exact Half-Space Proximal (HSP) neighbors of a query. The major emphasis of this approach is to minimize the number of distance computations, assuming distances and search time is correlated. While the implementation saves tremendously on distance computations, the search time is falling behind. By profiling the code, we can see where the search time is spent.
For the experiment, using a two-layer hierarchy on a dataset of $6D$ uniformly distributed points ($N=51,200$) with 10k
queries. Compiling with the flags -g, -pg, -O1 and the gprof2dot flags -s -w -n 0.5.
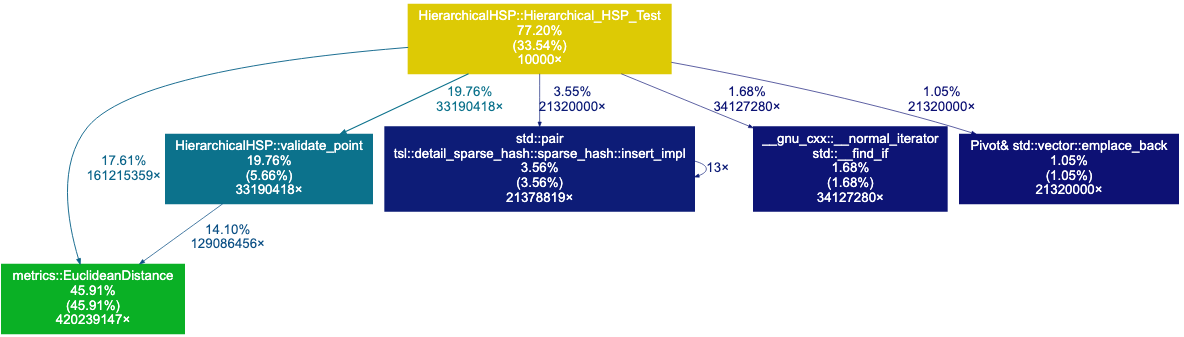
The original code gives us the following result: distances=20755.78, time(ms)=3.3016
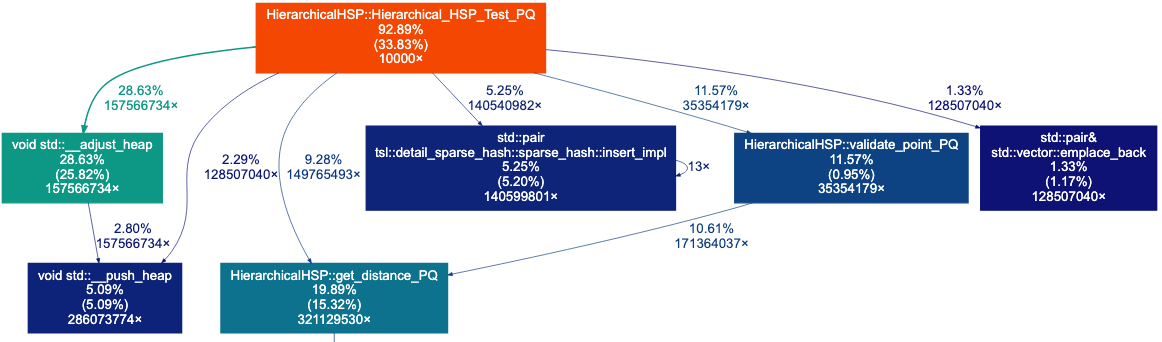
The Call Graph above shows that 28.63%+5.09% of the time is spent on the adjust_heap and push_heap functions, which are used by the priority queue. We also see that about 20% of the time is spent on distance computations. Let’s start by switching from a priority queue to a vector- we may not need to keep our points to be sorted by distance.
Now, we see a increase an distance and substantial decrease in time: 28810.75 distances, time(ms)=1.0722
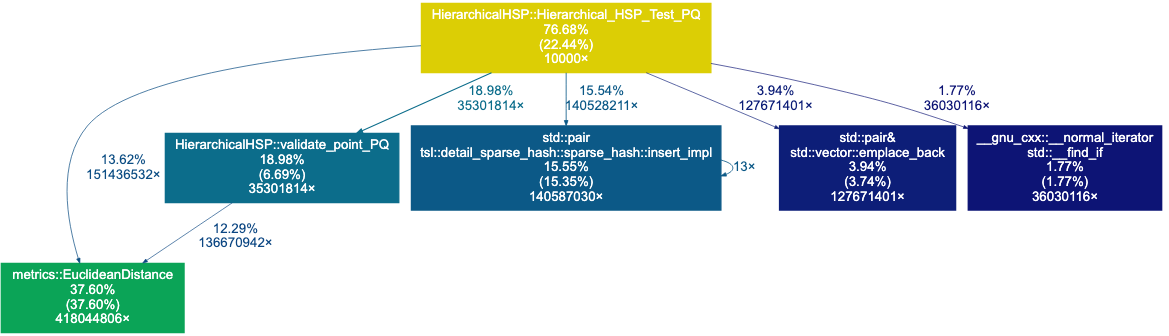
This iteration shows us that distance computations take a larger role. Now, we see that some std::pair tsl::detai_sparse...
data structure takes 15.55% of time. This is likely due to our frequent calls to the unordered map holding the radius
of each pivot. Let’s create a Struct that holds the <index,distance,radius,children> for each pivot instead
of requiring frequent fetches.
With some other adjustments too, we now have: 29030.18 distances and time(ms)=0.8179